Research Activities
The ”Computational Neuroimaging” (CN) group is headed by Kristoffer H. Madsen. The main focus of the group is modelling and analysis of neuroimaging data based on machine learning methodology. The efforts within the group aim to improve sensitivity and interpretability of the vast amounts of data that are acquired with neuroimaging techniques through sophisticated modelling and analysis methods. Furthermore, the group makes significant contributions to a wide range of projects running within DRCMR.
The CN group maintains particularly close collaboration with the “Neurophysics” group headed by Axel Thielscher, the “Biophysically Adjusted State-Informed Cortex Stimulation (BaSiCs)” group headed by Hartwig R. Siebner, and the “Computational Neuroscience of Reward” group headed by Oliver Hulme.
Key Research Areas
Modelling of functional connectivity
By continuous observation of brain activity, connectivity between brain areas can be inferred through the identification of statistical dependencies between signal from distinct brain areas. The research efforts within this area are mainly focused on unsupervised multivariate modelling 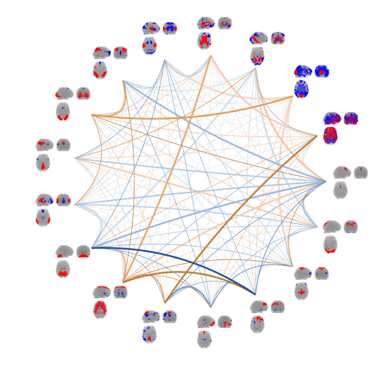 techniques for functional magnetic resonance imaging data (fMRI). Whereas traditional analyses of fMRI data typically involve inferring brain activity for each volume element (voxel) individually using a so-called mass-univariate approach. In contrast unsupervised multivariate modelling techniques aims to identify latent networks of functional connectivity by simultaneously considering all voxels in the brain in a combined multivariate model. Current efforts within this area are on multi-way extensions of unsupervised decomposition models, such as the well-known Independent Component Analysis, to naturally extend modelling of data recorded over several trials, sessions, subjects and modalities. One example is a Bayesian formulation of Independent Vector Analysis for modelling of functional networks across subjects in the presence of limited inter-subject variability. Additional research efforts are within Bayesian nonparametric modelling of fMRI data and investigations of predictability and reproducibility of unsupervised decomposition methods using resampling techniques.
techniques for functional magnetic resonance imaging data (fMRI). Whereas traditional analyses of fMRI data typically involve inferring brain activity for each volume element (voxel) individually using a so-called mass-univariate approach. In contrast unsupervised multivariate modelling techniques aims to identify latent networks of functional connectivity by simultaneously considering all voxels in the brain in a combined multivariate model. Current efforts within this area are on multi-way extensions of unsupervised decomposition models, such as the well-known Independent Component Analysis, to naturally extend modelling of data recorded over several trials, sessions, subjects and modalities. One example is a Bayesian formulation of Independent Vector Analysis for modelling of functional networks across subjects in the presence of limited inter-subject variability. Additional research efforts are within Bayesian nonparametric modelling of fMRI data and investigations of predictability and reproducibility of unsupervised decomposition methods using resampling techniques.
Source localization for EEG/MEG
EEG and MEG data records electrical and magnetic electrophysiological signals respectively over a limited number of electrodes or sensors (in the order of hundreds) outside the skull. The so-called source localization problem is concerned with identifying the origin/source of these signals inside the brain. This ill-posed problem is challenging mainly because the conductive properties of the head renders the signals measured outside the skull very correlated, and because the number of sensors available are typically much less that the potential number of locations for the sources. In this research we aim to investigate how the quality of the head model, accuracy of the conductive properties of the head and the source localization model type and regularization. This is approached through rigorous biophysical head modelling in collaboration with the neurophysics group and functional multimodal validation experiments involving fMRI and simultaneous EEG/MEG of well established behavioral paradigms.







